The cancer detectives
Albert Levan, a researcher at Lund University, was the first to establish how many chromosomes there are in the human genome. It was long believed that there were 48 chromosomes in total, but no one was sure because it was difficult to obtain good microscope slides. In the 1950s, Albert Levan and American Joe Hin Tjio improved the method, and in 1956 they established that the human genome is divided into 46 chromosomes, i.e. 23 pairs of chromosomes.
Felix Mitelman is another prominent researcher from Lund University. He has spent over 30 years studying various aspects of the changes to chromosomes that lead a cell to change and develop into a cancer cell, with a particular focus on leukaemia. Felix Mitelman began early on to compile these and other chromosome changes. All known chromosome changes in cancer have now been gathered in a large database, “The Mitelman database of chromosome aberrations”. Over 60 000 cases are described in the database. The results of the survey are that wide variation can be observed; almost all tumours have chromosome changes and some are typical of different types of cancer. Felix Mitelman’s initiative of mapping all chromosome aberrations in cancer has helped to improve diagnosis, treatment and prognosis for different types of cancer.
With the development of sequencing techniques, it is now possible to study the structure of the genome in detail, defined by the order of the nitrogenous bases (nucleotides) adenine, guanine, cytosine and thymine in the DNA. Using sequencing, the epigenome’s methyl groups have been studied, which is what enabled the twin studies that first demonstrated how our epigenome changes as we age. Detailed genetic and epigenetic sequencing now form the basis of the hunt by cancer researchers for diagnostically useful changes.
Text: Pia Romare
Published: 2014
Facts
-
Sequencing
-
Sequencing is a method used to decide the exact order of the building blocks of DNA or RNA. Sequencing of DNA gives a direct picture of our genetic make-up, while sequencing of the RNA that is naturally present in cells gives a picture of which parts of our non-coding DNA are used and in what way.
-
The image shows...
-
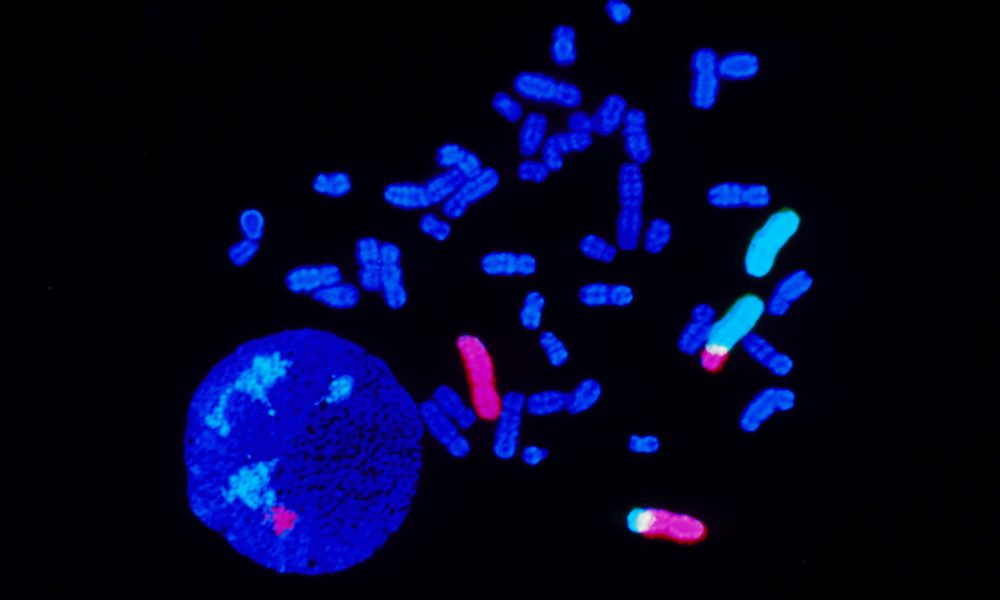
The image shows chromosomes from a cancer patient, showing 2:3 translocation. Fluorescence “in situ” hybridization (FISH) technique enables chromosomes to be tagged with fluorescent dyes to highlight genetic traits. Chromosomes are elongated, coming from the nucleus (round, lower left). Chromosomes 2 and 3 fluoresce pink and light blue; segments of these chromosomes have translocated so that part of chromosome 2 is attached to 3, and 3 to 2. Photographed at Cytocell Ltd., England.